Abstract
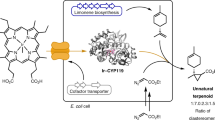
Nature has evolved an exquisite yet limited set of chemical reactions that underpin the function of all living organisms. By contrast, the field of synthetic organic chemistry can access reactivity not observed in nature, and integration of these abiotic reactions within living systems offers an elegant solution to the sustainable synthesis of many industrial chemicals from renewable feedstocks. Here we report a biocompatible Lossen rearrangement that is catalysed by phosphate in the bacterium Escherichia coli for the transformation of activated acyl hydroxamates to primary amine-containing metabolites in living cells. Through auxotroph rescue, we demonstrate how this new-to-nature reaction can be used to control microbial growth and chemistry by generating the essential metabolite para-aminobenzoic acid. The Lossen rearrangement substrate can also be synthesized from polyethylene terephthalate and applied to whole-cell biocatalytic reactions and fermentations generating industrial small molecules (including the drug paracetamol), paving the way for a general strategy to bioremediate and upcycle plastic waste in native and engineered biological systems.

Similar content being viewed by others
Main
The development of biocompatible reactions—non-enzymatic chemical transformations that can be interfaced with cellular metabolism—is a nascent approach to expanding the synthetic repertoire of living systems1,2,3,4,5. Using synthetic strategies established in modern organic chemistry, biocompatible reactions can be applied to the control of cellular function6,7,8,9, the diversification of metabolites in vivo10,11,12 and biological access to otherwise recalcitrant feedstocks for industrial biotechnology13,14,15. This approach complements existing methods for abiotic catalysis in cells16,17,18, including directed evolution19,20, the creation of non-native active sites using artificial cofactors or unnatural amino acids21, and the bottom-up design of enzymes using computational methods22. However, the reconstitution of new-to-nature biocatalysts in vivo is challenging and has often limited their application to in vitro reactions. The metabolic integration of non-native chemistries in living cells and particularly within the context of cellular metabolism remains a great challenge in the field of chemical biotechnology2.
Strategies to increase the limited toolbox of metabolic chemistry for microbial synthesis would enable the bioproduction of an increased range of industrial small molecules from sustainable feedstocks using engineered biology, lowering existing chemical manufacturing routes reliance on diminishing fossil fuels. Recent work in this area is limited to the use of artificial metalloenzymes in microbial cells that have been metabolically engineered to generate substrate(s) and apoenzyme, followed by transport of a non-native cofactor to the cell interior for biosynthesis23. To this end, Huang et al. reported the non-natural cyclopropanation of metabolically derived limonene in engineered Escherichia coli using an intracellular Ir-CYP119 metalloenzyme and exogenous ethyl diazoacetate24. Import of the Ir-porphyrin cofactor for biosynthesis was enabled by co-expressing the hug operon haem transport system from Plesiomonas shigelloides alongside a heterologous limonene biosynthetic pathway and together enabled the bioproduction of an unnatural cyclopropane-containing terpenoid in 0.25 mg l−1 and a 1:7.0:2.3:1.5 ratio of diastereoisomers. Biocompatible cyclopropanation chemistry has also been demonstrated at the outer membrane of E. coli and in membrane-associated micelles using a non-enzymatic Fe-phthalocyanine catalyst to intercept styrene generated in vivo from d-glucose via engineered metabolism (95% yield, 553 mg l−1)25. More recently, however, Huang et al. have achieved the complete integration of non-native carbene-transfer-based cyclopropanation chemistry into microbial metabolism in the bacterium Streptomyces albus J107426. Here, the authors combine a heterologous styrene biosynthesis pathway from L-phenylalanine with the native biosynthesis of the diazo-containing natural product azaserine and an Ir(Me)MPIX artificial metalloenzyme to enable the bioproduction of an unnatural cyclopropane-containing metabolite from entirely biobased building blocks in vivo (0.22 mg l−1). In addition to combining the end products of two metabolic pathways using an artificial enzyme, in a different report, Liu et al. used a hemin-catalysed oxidative decarboxylation reaction to convert metabolically generated α-acetolactate to diacetyl and subsequently (S,S)-2,3-butanediol in engineered Lactococcus lactis27. Most recently, Dennis et al. demonstrated the biocompatible organocatalytic α-methylenation of metabolically generated butyraldehyde, which was then subsequently reduced to 2-methylbutanal in engineered E. coli28. Together, these studies demonstrate the metabolic flexibility of microorganisms and how chemical principles can be used within metabolic pathways using abiotic catalysis to generate new-to-nature small molecules in cells.
An unexplored area of biocompatible chemistry is the non-enzymatic rearrangement of activated carboxylate substrates and their integration with native and engineered metabolic pathways in cells. Discovered in 1872 by Wilhelm Lossen, the Lossen rearrangement is characterized by the thermal- or metal-catalysed expulsion of a carboxylate from a bis-acylated hydroxylamine substrate29 (Fig. 1a). The rearrangement typically involves a phenyl hydroxamate ester and proceeds under basic conditions via 1,2-aryl migration to form an isocyanate that rapidly reacts with water under aqueous conditions to form a carbamic acid, followed by decarboxylation to the primary amine product30,31. The reaction is synthetically useful as the Lossen rearrangement substrate can be formed from readily available carboxylic acids, avoids the use of azide reagents (cf. the Curtius rearrangement) and occurs under mild conditions32,33. Overall, the reaction generates primary amines from carboxylate substrates with an accompanying one-carbon contraction, contrasting with the enzymatic chemistry enabled by ammonia lyases and aminotransferases34,35,36. Lossen-type rearrangements have been observed in vitro as unproductive substrates37 using chymotrypsin and are found as intermediates responsible for hydroxamic acid toxicity in Salmonella typhimurium TA9838. However, to the best of our knowledge, the Lossen rearrangement has never been interfaced with microbial metabolism for biocompatible chemistry and therefore remains a functional group transformation that is unique to the field of synthetic organic chemistry.
a, A comparison of strategies for C–N bond formation via Lossen rearrangement in synthetic organic chemistry or via chorismate pathways in cellular metabolism. b, The proposed merging of non-enzymatic Lossen rearrangement chemistry with cellular metabolism for sustainable synthesis and the bio-upcycling of plastic waste. LG, leaving group.
Here, we report the biocompatible Lossen rearrangement of acyl hydroxamates in living cells and interface this abiotic reaction with cellular metabolism for native and de novo biosynthesis in E. coli. The reaction occurs in vivo, under ambient conditions, is non-toxic to E. coli and is catalysed by phosphate in cells. We go on to synthesize the Lossen rearrangement substrate from polyethylene terephthalate (PET) and show through auxotroph rescue experiments how E. coli growth and metabolism can be dependent upon a plastic-bottle-derived small molecule (Fig. 1b). Finally, metabolic cooperation with the Lossen rearrangement is demonstrated through the conditional biotransformation of exogenous alkenes and of PET-derived para-aminobenzoate (PABA) to the analgesic and antipyretic drug paracetamol (para-hydroxyacetanilide). Overall, this work expands the available toolbox of metabolic chemistry for small-molecule synthesis in native and engineered cells.
Results and discussion
Reaction screening via auxotroph rescue
Inspired by Li et al. on the Fe-phenanthroline-catalysed Lossen rearrangement of heteroauxin-derived substrates in dichloromethane at room temperature and the reported hemin-catalysed N–O insertion chemistry in artificial enzymes39,40,41,42, we reasoned that a biocompatible metal catalyst would be effective in generating an intermediate nitrenoid from an O-pivaloyl (O-Piv)-substituted benzhydroxamate substrate under aqueous conditions39. We therefore set out to investigate whether the Lossen rearrangement was biocompatible and whether it could be interfaced with microbial metabolism to enable non-natural biosynthesis. To test catalyst reactivity and biocompatibility simultaneously, we designed an auxotroph rescue experiment where a para-carboxyl O-pivalolylhydroxamate Lossen rearrangement substrate 1 would generate the essential metabolite PABA in situ. PABA is essential for folic acid biosynthesis in bacteria, and organisms deficient in PABA cannot grow due to defects in nucleotide and DNA metabolism6,43. Auxotrophic organisms (including humans) must therefore sequester these essential nutrients from their surrounding environment or from other (micro)organisms living in close proximity. Therefore, a successful Lossen rearrangement of 1 would generate PABA and result in growth of the auxotrophic cells that could be detected by optical density (OD600) measurement (Fig. 2a). Moreover, toxic catalysts would inhibit cell growth irrespective of PABA generation and, therefore, successful growth would be a positive indicator of both catalyst activity and biocompatibility. To this end, O-Piv benzhydroxamate 1 was synthesized in two steps from 4-formylbenzoic acid via amide bond formation with O-Piv hydroxylamine followed by aldehyde oxidation using periodic acid and catalytic pyridinium chlorochromate (Supplementary Scheme 1). We selected a series of K-12-derived Keio collection knockout strains including E. coli BW25113∆pabB deficient in the catalytic subunit of aminodeoxychorismate synthase that is involved in the penultimate step of PABA biosynthesis (Fig. 1a and Supplementary Fig. 1). The catalyst screen was populated with metal complexes that have been reported in the literature to promote the Lossen or Curtius rearrangement and/or analogous reactions under mild or aqueous reaction conditions39,44,45. These include FeCl2, hemin, iron phthalocyanine, ferroin, ZnTPP, Zn(PPIX) and Zn(acac)2. Lossen substrate 1 (10 µM) was added to growth tubes containing M9-glycerol medium and catalyst (10 mol%) followed by E. coli BW25113∆pabB inoculated from a saturated starter culture grown in M9-glycerol medium containing PABA (105 dilution). Cultures were then incubated for 72 h at 37 °C at 220 r.p.m. (Fig. 2a). As expected, no cell growth was observed in the absence of PABA. However, we were surprised to observe that cell growth occurred in every other tube (Fig. 2b,c). Crucially, growth was observed in control cultures containing 1 and no catalyst, indicating that the Lossen rearrangement of 1 was biocompatible and either occurring spontaneously in growth media, being promoted by cellular components (for example, membranes) or being catalysed by a native enzyme in E. coli (Fig. 2d,e). To eliminate the latter two, PABA was quantified using a N-(1-napthyl)ethylenediamine colorimetric assay in control reactions performed in M9-glycerol only (Fig. 2f and Supplementary Fig. 2). Indeed, PABA was detected in the absence of cells and was not detected when 1 was incubated in ultrapure H2O. Sequential elimination of each component of M9 medium (NH4Cl, CaCl2, MgSO4 and HPO42−) and comparison with PABA generation in phosphate-buffered saline (PBS) revealed that the Lossen rearrangement of 1 was mediated by phosphate. Aniline formation from N-methyl O-acetyl benzhydroxamic acid 3 or benzhydroxamic acid 4 was also abolished under these conditions (Fig. 2g and Supplementary Fig. 3), demonstrating that the reaction probably proceeds via initial N–H deprotonation followed by 1,2-aryl migration, rather than ester hydrolysis followed by a phosphate-catalysed rearrangement of the corresponding hydroxamic acid (Fig. 2g). Spectrophotometric analysis of the cultures revealed that the highest OD600 was observed in cultures incubated in the presence of 1 and either FeCl2, ferroin or Fe(acac)3 (Fig. 2c). Intriguingly, these complexes are the weakest Fe binders, and therefore the higher growth of these cultures is probably due to a non-enzymatic phosphate-catalysed Lossen rearrangement followed by increased Fe availability to the microorganism during the log-phase of growth, which is limiting in the tested conditions. Auxotroph rescue was also confirmed in Keio knockout strains E. coli BW25113∆pabA and E. coli BW25113∆aroC, and both displayed either an extended lag phase or lower final cell density when compared with E. coli BW25113∆pabB grown in the presence of PABA or 1 (Supplementary Fig. 1). The toxicity of 1 to E. coli BW25113∆pabB was determined by a serial dilution and plate count assay. All substrates were biocompatible with E. coli growth by OD600 and colony-forming units (CFU) per millilitre in the 10–1,000 µM range (Fig. 2e and Supplementary Fig. 4). A panel of O-acyl substrates containing hydrophilic and hydrophobic groups were screened and converted to aromatic amines with no notable rate increase by colorimetric assay, except for a rapid product formation when using pentafluorobenzyl substrate S2 (Supplementary Fig. 3) and abolished Lossen reactivity using the hydrophilic O-succinyl substrate S6 (Supplementary Fig. 3). Competing hydrolysis to benzhydroxamic acid 4 was observed for the O-Ac substrate and so 1 was selected for further study. Finally, high-performance liquid chromatography (HPLC) quantification over time in the presence and absence of replicating E. coli BW25113∆pabB cells revealed increased substrate consumption after >24 h at the point where cells enter log-phase growth, indicating that the Lossen rearrangement of 1 is not only biocompatible but also potentially accelerated in the presence of metabolically active cells (Fig. 2h).
a, Auxotroph rescue experiment via Lossen rearrangement. b, A photograph of cultures after the auxotroph recovery of E. coli BW25113∆pabB in the presence of 1 and various transition-metal catalysts. c, Culture density measurements by spectrophotometry. d, Auxotroph rescue growth curves from cultures of E. coli BW25113∆pabB grown in the presence of PABA or 1. The extended lag phase is due to the 105-fold dilution of the starting inoculum that is required to ensure no background growth without PABA. e, Plate-count assay from cultures of E. coli BW25113∆pabB grown in the presence of PABA or 1. f, In vitro experiment examining the effect of M9 media components on the Lossen rearrangement of 1. g, The proposed phosphate-catalysed Lossen rearrangement of 1 to PABA and in vitro reactivity of O-acylated and N-methylated control compounds 2 and 3. h, Substrate depletion assay using 200 µM 1 in the presence and absence of metabolically active cells. ***P < 0.0005 (t-test). All data are presented as mean values ± s.d. of three biological replicates.
PET bioremediation
Despite our initial focus on developing biocompatible reactions for synthesis in microbial cells, this intriguing observation suggested a method for the bioremediation of 1 derived from waste materials. For example, the synthesis of 1 can be envisioned from terephthalic acid—the depolymerization monomer of PET plastic waste. We therefore reasoned that we could synthesize 1 from PET and by introducing PABA auxotrophy into a microbial strain effectively condition cell growth to the presence of PET-derived small molecules, offering a strategy for the remediation of this prolific waste material and environmental pollutant into microbial biomass. Industrial PET production worldwide is currently 56 million tons per year, and approximately 80% of this is designed to be single-use, leading to around 24 million tons of PET waste each year that is either incinerated or sent to landfill46. To this end, a synthesis of 1 was optimized from a discarded plastic bottle in two steps via the hydrolysis of PET flakes to terephthalic acid, followed by amide coupling with O-Piv hydroxylammonium triflate and propylphosphonic anhydride (T3P) to generate PET-1 (Fig. 3a and Supplementary Scheme 1). Auxotroph recovery and growth of E. coli BW25113∆pabB was observed using this PET-derived substrate, accompanied by quantitative reduction in substrate concentration and no detectable PABA after 48 h (Fig. 3b–d). Growth in the presence of PET-1 had a comparable rate (0.25 h−1 for PABA and 0.33 h−1 for PET-1 after 40 h), final cell density (OD600) and viability count (CFU ml−1) at stationary phase and led to a similar decrease in substrate compared with using 1 (Fig. 3d and Supplementary Fig. 5).
a, Synthesis of the Lossen rearrangement substrate from PET waste and PET bioremediation strategy via auxotroph recovery. b, Growth curves from 96-well plate experiments during auxotroph rescue. Error bars are omitted for clarity. c, A photograph of auxotroph rescue experiments using a PET-derived substrate. d, Substrate depletion (grey) and dry cell weight (d.c.w.) production (blue) during growth experiments in Falcon tubes using 1 µM PET-1. A conversion factor of 0.33 grams per litre per OD600 was applied. All data are shown as triplicate experiments to one standard deviation. (i) NaOH, EtOH/H2O, reflux, 10 h. (ii) T3P, iPr2NEt, H2NOPiv·TfOH, tetrahydrofuran, 0 °C to room temperature, 16 h.
Interfaced metabolic synthesis
Having confirmed that the Lossen rearrangement was biocompatible and could be used to bioremediate PET-derived substrates, we next moved on to examine whether rescued cells could be used for biocatalytic reactions (Fig. 4a). We decided to focus this study initially on the whole-cell C=C bond reduction of maleates and keto-acrylates reported by Brewster et al. under fermentation conditions using native reductases in E. coli47. The substrates dimethyl maleate (DMM, 5) and keto-acrylates (E)-7 and (Z)-7 were synthesized and added to cultures of E. coli BW25113∆pabB grown in the presence of 1. Pleasingly, all the substrates were quantitatively reduced to dimethyl succinate 6 or γ-ketoester 8 by 1H nuclear magnetic resonance (NMR) after 24 h at 37 °C and product formation was observed only in the presence of PABA or 1, indicating that the biocompatible Lossen rearrangement could be used to control the chemical output of a microbial fermentation (Fig. 4b and Supplementary Figs. 6 and 7). Having interfaced the Lossen rearrangement with cell growth, metabolism and native biosynthetic reactions, we moved on to assess whether the Lossen rearrangement product could be syphoned into a de novo metabolic pathway in vivo. To test this, we decided to target the biosynthesis of paracetamol (para-hydroxyacetanilide) 10 from 1 in engineered E. coli. Both 1 and 10 are non-toxic to E. coli BW25113∆pabB at concentrations up to 1 mM (265 mg l−1; 1) and 6.6 mM (1 g l−1; 10) (Supplementary Fig. 8). Paracetamol is the first-in-line World Health Organization-recommended treatment for pain and fever worldwide48. It is currently manufactured from phenol (derived from fossil fuels by the cumeme process) via nitration, reduction and N-acetylation with acetic anhydride before being formulated into an orally available medication49. By contrast, 10 can be synthesized in cells from PABA via 4-aminophenol (4-AP, 9) by two enzymes: (1) an O2- and NADH-dependent aminobenzoate hydroxylase (ABH60) from the fungus Agaricus bisporus and (2) a K211G mutant of the acetyl CoA-dependent arylamine N-acyltransferase from the bacterium Pseudomonas aeruginosa (PANAT)50 (Fig. 4c). Paracetamol biosynthesis has been reported from d-glucose in E. coli but not from a waste feedstock or a PET-derived substrate. To this end, the genes encoding ABH60 and PANAT were synthesized and cloned into a Joint Universal Modular Plasmid (JUMP) vector generating plasmid pSWL112 (Supplementary Fig. 9 and Supplementary Table 3). The kanamycin resistance cassette from E. coli BW25113∆pabB Keio strain was removed using a pCP20-encoded flippase before transformation with pSWL112, generating an engineered and auxotrophic strain to produce paracetamol that could be recovered using 1 (Supplementary Fig. 10a). However, HPLC analysis of cultures of E. coli BW25113∆pabB_pSWL112 grown in the presence of 1 identified 4-acetamidobenzoic acid (4-AB, 31 mg l−1) as the sole product from the paracetamol pathway, resulting from unproductive N-acetylation of PABA by PANAT and acetyl-CoA (Fig. 4c and Supplementary Fig. 10b). To overcome this, a series of modified JUMP plasmids were designed containing abh60 and panat genes under the control of constitutive and inducible promoters to optimize flux from PABA to paracetamol. Two optimization strategies were trialled: (1) growth of E. coli BW25113∆pabB_pSWL156_ pSWL354 or E. coli BW25113∆pabB_pSWL156_ pSWL355 in the presence of 1 with constitutive expression of abh60 (pSWL156, J23100-abh60) and panat expression in plasmids pSWL354 (/Pbad-panat) and pSWL355 (/Ptet-panat) induced after 72 h (Supplementary Figs. 10b and 12); or (2) the separation of ABH60- and PANAT-catalysed reactions using 2 E. coli strains (BW25113∆pabB_pSWL156 or BW25113∆pabB_pSWL350, with BL21(DE3)_pSWL157 (/T7IPTG-panat)) (Supplementary Fig. 13) by the addition of pre-expressed E. coli BL21(DE3)_pSWL157 cells (OD600 of 20) after growth of the PABA auxotroph expressing ABH60 in the presence of 1. Both approaches eliminated 4-AB production and generated paracetamol in 12 mg l−1 (Supplementary Fig. 10b) and 19 mg l−1 (29% yield), respectively, from 1 (Fig. 4c and Supplementary Fig. 14; see Supplementary Figs. 15 and 16 for a comparison with different promoter types and control reactions). To enhance the synthetic utility of paracetamol synthesis from 1 and PET-1, a one-pot two-step procedure was optimized where the Lossen rearrangement was first initiated at 50 °C in aqueous phosphate buffer (200 mM, 50 °C, pH 8.0) followed by addition of induced E. coli BW25113ΔpabB_p354 and E. coli BW25113ΔpabB_p350 whole cells expressing panat and abh60, respectively (1:200, 37 °C, OD600 of 12.5–25). Under these conditions, quantitative yield of paracetamol was observed from PABA and in 86% from 1 (Fig. 4d). Using the plastic-waste-derived substrate PET-1 under analogous biotransformation conditions afforded paracetamol in 83% yield. Finally, reducing the arabinose concentration during protein expression enabled a reduction in the ratio of panat and abh60 expressing strains to 1:100 and increased the final yield of paracetamol 10 to 92% from PET-1 (Fig. 4d). Intensification of this process will focus on in situ biocatalytic depolymerization of industrial PET samples for both bioremediation and bio-upcycling at bioreactor scale to further improve overall productivity and product isolation. This will be accompanied by quantitative sustainability analyses via life cycle assessment to ensure process optimizations achieve maximum environmental sustainability gains. Future work will also focus on further optimization of the de novo pathway to maximize flux into paracetamol biosynthesis using synthetic biology approaches as well as applying the biocompatible Lossen rearrangement to other chemo-enzymatic cascades and fully integrating this new-to-nature reaction within metabolically evolved microorganisms.
a, Interfacing the Lossen rearrangement with native and engineered biosynthetic pathways in E. coli. b, Biotransformation of DMM (5) to dimethylsuccinate (6) and β-ketoacrylate 7 to γ-ketoester 8 using E. coli BW25113∆pabB and 1 or PET-1. c, A de novo biosynthetic pathway to 4-AP (9) and paracetamol 10 incorporating a non-enzymatic Lossen rearrangement, plasmid designs and whole-cell production experiments. d, Paracetamol synthesis by one-pot Lossen rearrangement and bacterial whole-cell synthesis. Strain E. coli_p350 expresses ABH60 and strain E. coli_p354 expresses PANATK211G. Ratios refer to E. coli_p354 and E. coli_p350 in biotransformations (1:1, OD600 26; 1:5, OD600 16; 1:100 and 1:200, OD600 12.5). aFinal cell density OD600 25. OD refers to the final optical density at 600 nm. bCells induced with 0.5% l-arabinose. Reaction conditions: (i) substrate (0.5 mM), aqueous potassium phosphate (200 mM, pH 8.0), 50 °C, 48 h; (ii) E. coli_p354 and E. coli_p350 (1:10; OD600 20). Biotransformations were analysed by 1H NMR relative to an internal standard of trimethoxybenzene or by HPLC relative to an internal standard of caffeine. All data are presented as mean values ± s.d. of three biological replicates.
Conclusions
This study reports the discovery of a biocompatible Lossen rearrangement that can be interfaced with cellular metabolism in the bacterium E. coli. The non-enzymatic reaction proceeds in the presence of bacterial cells, is non-toxic and is catalysed by mono- or di-basic phosphate at neutral pH, highlighting a multifaceted role of phosphate ions in living cells for pH homeostasis, membrane biosynthesis and now biocompatible non-enzymatic chemistry. The biocompatible reaction also forms primary amine products in vivo via a mechanism that is distinct from known biosynthetic logic and, therefore, provides a useful tool in metabolic engineering for the generation of amine-containing metabolites. We demonstrate through auxotroph rescue how PABA can be generated by the Lossen rearrangement in vivo and used to control microbial growth and chemistry in fermentation and whole-cell reactions. Synthesis of the Lossen rearrangement substrate was achieved from a waste PET bottle and incorporated metabolically to generate biomass and control whole-cell biotransformations. The substrate was syphoned into a de novo biosynthetic pathway to paracetamol, demonstrating the production of this essential medication from plastic waste via a strategy that cannot be achieved using chemical synthesis or biological synthesis alone. Biocompatible chemistry should therefore be considered as complementary to nascent work in enzyme design and engineering for abiotic chemistry and integrated cooperatively as a tool in living cells to expand the synthetic chemistry that is possible within engineered biological systems.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available in the article and its Supplementary Information. Source data are provided with this paper.
References
Wallace, S., Schultz, E. E. & Balskus, E. P. Using non-enzymatic chemistry to influence microbial metabolism. Curr. Opin. Chem. Biol. 25, 71–79 (2015).
Sadler, J. C., Dennis, J. A., Johnson, N. W. & Wallace, S. Interfacing non-enzymatic catalysis with living microorganisms. RSC Chem. Biol. 2, 1073–1083 (2021).
Wallace, S. & Balskus, E. P. Opportunities for merging chemical and biological synthesis. Curr. Opin. Biotechnol. 30, 1–8 (2014).
Stewart, K. N. & Domaille, D. W. Enhancing biosynthesis and manipulating flux in whole cells with abiotic catalysis. ChemBioChem 22, 469–477 (2021).
Ngo, A. H., Bose, S. & Do, L. H. Intracellular chemistry: integrating molecular inorganic catalysts with living systems. Chemistry 24, 10584–10594 (2018).
Lee, Y., Umeano, A. & Balskus, E. P. Rescuing auxotrophic microorganisms with nonenzymatic chemistry. Angew. Chem. Int. Ed. 52, 11800–11803 (2013).
Fan, G., Dundas, C. M., Graham, A. J., Lynd, N. A. & Keitz, B. K. Shewanella oneidensis as a living electrode for controlled radical polymerization. Proc. Natl Acad. Sci. USA 115, 4559–4564 (2018).
Guo, J. et al. Light-driven fine chemical production in yeast biohybrids. Science 362, 813–816 (2018).
Rubini, R., Ivanov, I. & Mayer, C. A screening platform to identify and tailor biocompatible small-molecule catalysts. Chemistry 25, 16017–16021 (2019).
Wallace, S. & Balskus, E. P. Interfacing microbial styrene production with a biocompatible cyclopropanation reaction. Angew. Chem. Int. Ed. 54, 7106–7109 (2015).
Maaskant, R. V., Chordia, S. & Roelfes, G. Merging whole-cell biosynthesis of styrene and transition-metal catalyzed derivatization reactions. ChemCatChem 13, 1607–1613 (2021).
Sharma, S. V. et al. Living GenoChemetics by hyphenating synthetic biology and synthetic chemistry in vivo. Nat. Commun. 8, 229 (2017).
Valenzuela-Ortega, M., Suitor, J. T., White, M. F. M., Hinchcliffe, T. & Wallace, S. Microbial upcycling of waste PET to adipic acid. ACS Cent. Sci. 9, 2057–2063 (2023).
Dennis, J. A., Sadler, J. C. & Wallace, S. Tyramine derivatives catalyze the aldol dimerization of butyraldehyde in the presence of Escherichia coli. ChemBioChem 23, e202200238 (2022).
Wu, S., Zhou, Y., Gerngross, D., Jeschek, M. & Ward, T. R. Chemo-enzymatic cascades to produce cycloalkenes from bio-based resources. Nat. Commun. 10, 5060 (2019).
Adamson, C. & Kanai, M. Integrating abiotic chemical catalysis and enzymatic catalysis in living cells. Org. Biomol. Chem. 19, 37–45 (2021).
Fu, Q. et al. Bioorthogonal chemistry for prodrug activation in vivo. Chem. Soc. Rev. 52, 7737–7772 (2023).
Rebelein, J. G. & Ward, T. R. In vivo catalyzed new-to-nature reactions. Curr. Opin. Biotechnol. 53, 106–114 (2018).
Arnold, F. H. Directed evolution: bringing new chemistry to life. Angew. Chem. Int. Ed. 57, 4143–4148 (2018).
Wang, Y. et al. Directed evolution: methodologies and applications. Chem. Rev. 121, 12384–12444 (2021).
Lechner, H. & Oberdorfer, G. Derivatives of natural organocatalytic cofactors and artificial organocatalytic cofactors as catalysts in enzymes. ChemBioChem 23, e202100599 (2022).
Meinen, B. A. & Bahl, C. D. Breakthroughs in computational design methods open up new frontiers for de novo protein engineering. Protein Eng. Design Select. 34, gzab007 (2021).
Davis, H. J. & Ward, T. R. Artificial metalloenzymes: challenges and opportunities. ACS Cent. Sci. 5, 1120–1136 (2019).
Huang, J. et al. Unnatural biosynthesis by an engineered microorganism with heterologously expressed natural enzymes and an artificial metalloenzyme. Nat. Chem. 13, 1186–1191 (2021).
Wallace, S. & Balskus, E. P. Designer micelles accelerate flux through engineered metabolism in E. coli and support biocompatible chemistry. Angew. Chem. Int. Ed. 55, 6023–6027 (2016).
Huang, J. et al. Complete integration of carbene-transfer chemistry into biosynthesis. Nature 617, 403–408 (2023).
Liu, J. et al. Combining metabolic engineering and biocompatible chemistry for high-yield production of homo-diacetyl and homo-(S,S)-2,3-butanediol. Metab. Eng. 36, 57–67 (2016).
Dennis, J. A., Johnson, N. W., Thorpe, T. W. & Wallace, S. Biocompatible α-methylenation of metabolic butyraldehyde in living bacteria. Angew. Chem. Int. Ed. 62, e202306347 (2023).
Lossen, W. Ueber Benzoylderivate des Hydroxylamins. Justus Liebigs Ann. Chem. 161, 347–362 (1872).
Thomas, M. et al. The Lossen rearrangement from free hydroxamic acids. Org. Biomol. Chem. 17, 5420–5427 (2019).
Ghosh, A. K., Sarkar, A. & Brindisi, M. The Curtius rearrangement: mechanistic insight and recent applications in natural product syntheses. Org. Biomol. Chem. 16, 2006–2027 (2018).
Bauer, L. & Exner, O. The chemistry of hydroxamic acids and N‐hydroxyimides. Angew. Chem. Int. Ed. 13, 376–384 (1974).
Citarella, A., Moi, D., Pinzi, L., Bonanni, D. & Rastelli, G. Hydroxamic acid derivatives: from synthetic strategies to medicinal chemistry applications. ACS Omega 6, 21843–21849 (2021).
Citoler, J., Derrington, S. R., Galman, J. L., Bevinakatti, H. & Turner, N. J. A biocatalytic cascade for the conversion of fatty acids to fatty amines. Green Chem. 21, 4932–4935 (2019).
France, S. P. et al. One-pot cascade synthesis of mono- and disubstituted piperidines and pyrrolidines using carboxylic acid reductase (CAR), ω-transaminase (ω-TA), and imine reductase (IRED) biocatalysts. ACS Catal. 6, 3753–3759 (2016).
Weise, N. J. et al. Bi‐enzymatic conversion of cinnamic acids to 2‐arylethylamines. ChemCatChem 12, 995–998 (2020).
Groutas, W. C., Giri, P. K., Crowley, J. P., Castrisos, J. C. & Brubaker, M. J. The Lossen rearrangement in biological systems. Inactivation of leukocyte elastase and alpha-chymotrypsin by (dl)-3-benzyl-N- (methanesulfonyloxy) succinimide. Biochem. Biophys. Res. Commun. 141, 741–748 (1986).
Lee, M. S. & Isobe, M. Metabolic activation of the potent mutagen, 2-naphthohydroxamic acid, in Salmonella typhimurium TA98. Cancer Res. 50, 4300–4307 (1990).
Li, D., Wu, T., Liang, K. & Xia, C. Curtius-like rearrangement of an iron–nitrenoid complex and application in biomimetic synthesis of bisindolylmethanes. Org. Lett. 18, 2228–2231 (2016).
Cho, I. et al. Enantioselective aminohydroxylation of styrenyl olefins catalyzed by an engineered hemoprotein. Angew. Chem. Int. Ed. 58, 3138–3142 (2019).
Jia, Z. J., Gao, S. & Arnold, F. H. Enzymatic primary amination of benzylic and allylic C(sp3)–H bonds. J. Am. Chem. Soc. 142, 10279–10283 (2020).
Athavale, S. V. et al. Biocatalytic, intermolecular C–H bond functionalization for the synthesis of enantioenriched amides. Angew. Chem. Int. Ed. 60, 24864–24869 (2021).
Wegkamp, A., Van Oorschot, W., De Vos, W. M. & Smid, E. J. Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Appl. Environ. Microbiol. 73, 2673–2681 (2007).
Lebel, H. & Leogane, O. Boc-protected amines via a mild and efficient one-pot curtius rearrangement. Org. Lett. 7, 4107–4110 (2005).
Kweon, J. & Chang, S. Highly robust iron catalyst system for intramolecular C(sp3)–H amidation leading to γ-lactams. Angew. Chem. Int. Ed. 60, 2909–2914 (2021).
Soong, Y. H. V., Sobkowicz, M. J. & Xie, D. Recent advances in biological recycling of polyethylene terephthalate (PET) plastic wastes. Bioengineering 9, 98 (2022).
Brewster, R. C., Suitor, J. T., Bennett, A. W. & Wallace, S. Transition metal‐free reduction of activated alkenes using a living microorganism. Angew. Chem. 131, 12539–12544 (2019).
Freo, U., Ruocco, C., Valerio, A., Scagnol, I. & Nisoli, E. Paracetamol: a review of guideline recommendations. J. Clin. Med. 10, 3420 (2021).
Friderichs, E., Christoph, T. & Buschmann, H. in Ullmann’s Encyclopedia of Industrial Chemistry 8–9 (John Wiley & Sons, 2007); https://doi.org/10.1002/14356007.a02_269.pub2
Hou, F., Xian, M. & Huang, W. De novo biosynthesis and whole-cell catalytic production of paracetamol on a gram scale in Escherichia coli. Green Chem. 23, 8280–8289 (2021).
Acknowledgements
We acknowledge an iCASE studentship EP/T517501/1 (N.W.J.) from AstraZeneca and EPSRC, a Future Leaders Fellowship MR/S033882/1 (S.W.) from UKRI and a Sustainable Manufacturing grant EP/W019000/1 (S.W.) from EPSRC. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
N.W.J. performed chemical synthesis, auxotroph rescue and biocatalysis reactions. M.V.-O., A.K. and N.W.J. performed cloning experiments. N.W.J., T.W.T. and Y.E. performed metabolite analysis experiments. The paper was written through contributions from all authors, and all authors have given approval to the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
N.W.J., M.V.-O., T.W.T., Y.E., A.K. and S.W. declare no competing interests. K.M. is an employee of AstraZeneca and may own stock options.
Peer review
Peer review information
Nature Chemistry thanks Jay Keasling and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Materials and methods, Supplementary Figs. 1–16, Tables 1–5 and Scheme 1, and NMR spectroscopy data.
Source data
Source Data Fig. 2
Raw data for Fig. 2.
Source Data Fig. 3
Raw data for Fig. 3.
Source Data Fig. 4
Raw data for Fig. 4.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Johnson, N.W., Valenzuela-Ortega, M., Thorpe, T.W. et al. A biocompatible Lossen rearrangement in Escherichia coli. Nat. Chem. 17, 1020–1026 (2025). https://doi.org/10.1038/s41557-025-01845-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-025-01845-5
This article is cited by
-
Everyday painkiller made from plastic — by E. coli
Nature (2025)
-
New-to-nature biocompatible chemistry for plastic waste upcycling
Nature Chemistry (2025)